Invasive species are a worldwide threat that alter ecosystems, endanger species, and cause immense economic burdens through their effects on human health and agriculture. While there have been extensive efforts to understand traits underlying invasibility and study the consequences of invasions across communities, food webs, and ecosystems, there is less consensus describing the evolutionary processes underlying invasions. Early hypotheses suggested that introduced populations were small, gene flow with the native range limited, and little selection acted during the invasion process – conclusions that have been largely overturned in the last decade. However, more nuanced insights into the process are still lacking: How much do initial colonization dynamics influence invasion trajectory? Does admixture play an important role in creating novel phenotypic variation? How much does selection influence establishment and spread? These questions are the focus of our work in the Kooyers lab and Consortium for Plant Invasion Genomics (or CPING).
Figure (left):The Consortium for Plant INvasion Genomics (CPING) is a network of collaborators dedicated to using genomics and bioinformatics tools to better understand the process of invasions. While the original network consisted of six collaborators using genomics in combination with herbarium specimens and contemporary collections to examine colonization, admixture and adaptation during invasions, we have expanded to include many more species and collaborators. You can find out more about CPING at our website: www.invasiongenomics.com
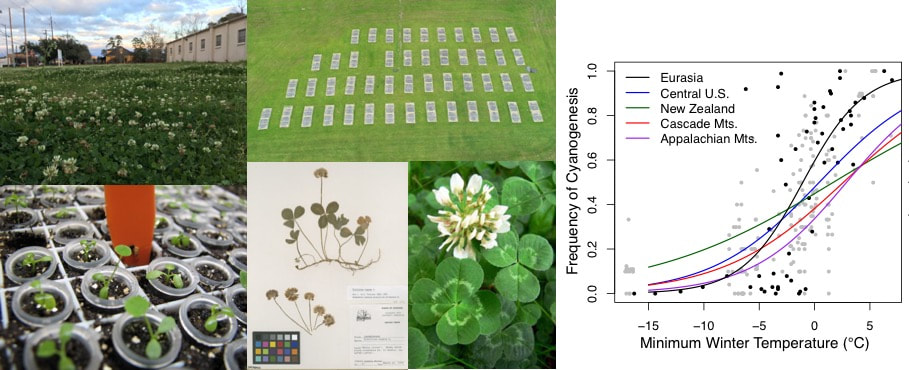
The Kooyers lab uses white clover (Trifolium repens) as a model system for understanding the role of rapid adaptation in facilitating invasions following introduction. White clover is native to Eurasia but was introduced as a forage legume around the world in the 18th-19th centuries. It can now be found invading feral fields, lawns, and grasslands on six continents. Across white clover's native and invasive ranges, it has repeatedly evolved clines in cyanogenesis, an important chemical defense in which a plant produces hydrogen cyanide in response to herbivory. Notably, white clover populations in warmer regions of both its native and invasive ranges have higher frequencies of cyanogenic individuals. The evolution of recurrent cyanogenesis clines suggests that rapid adaptation following introduction is common in white clover, making it an ideal system to evaluate how colonization, admixture, and selection interact during invasion.
We also have begun to expand our work beyond the cyanogenesis polymorphism into understanding the ecological genomics of adaptation during invasions. In collaborations with CPING collaborators, Ken Olsen at Wash U., and Marc Johnson, James Santangelo, and Lucas Albano at University of Toronto, Mississauga, we have an ambitious research program that incorporates manipulative experiments in the field and greenhouse, population genomics of contemporary populations and herbarium specimens, and a four-way transcontinental experimental garden to examine patterns of adaptation following introduction.
We have begun to collaborate with colleagues at different universities that are interested in asking similar questions with their organisms of interest. Please contact Nic if you are interested in getting involved in CPING or collaborating!
We also have begun to expand our work beyond the cyanogenesis polymorphism into understanding the ecological genomics of adaptation during invasions. In collaborations with CPING collaborators, Ken Olsen at Wash U., and Marc Johnson, James Santangelo, and Lucas Albano at University of Toronto, Mississauga, we have an ambitious research program that incorporates manipulative experiments in the field and greenhouse, population genomics of contemporary populations and herbarium specimens, and a four-way transcontinental experimental garden to examine patterns of adaptation following introduction.
We have begun to collaborate with colleagues at different universities that are interested in asking similar questions with their organisms of interest. Please contact Nic if you are interested in getting involved in CPING or collaborating!
Figure (clockwise from top left): White clover 'natural' population in Lafayette, Louisiana. Aerial view of field manipulative experiment at UL Ecology Center. Cyanogenesis clines on multiple continents. Herbivore preference trials. Herbarium sheets of white clover used within CPING temporal sampling design. A charismatic beauty - Trifolium repens